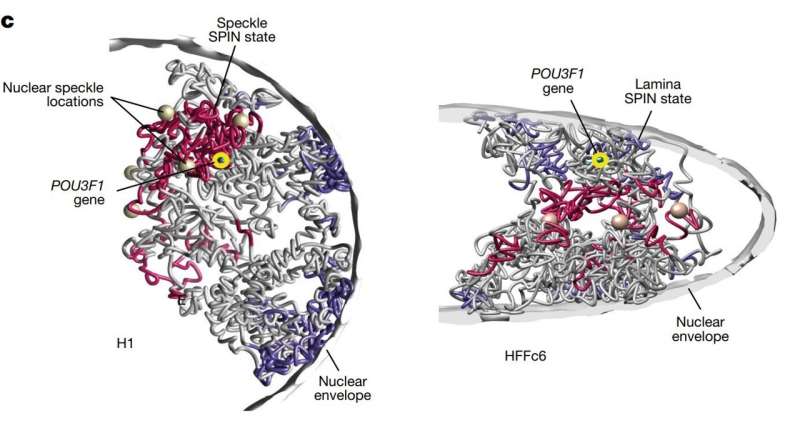
In a groundbreaking study, researchers have successfully charted more than 140,000 DNA loops within human cells, providing the most detailed map yet of how chromosomes are organized inside the nucleus. This achievement sheds new light on the three-dimensional architecture of the genome, with implications for understanding gene regulation, disease mechanisms, and potential future therapies.
DNA is not simply a linear sequence of nucleotides; it folds and loops inside the nucleus to regulate which genes are active and when. These loops allow distant regions of the genome to come into close physical contact, facilitating interactions that are critical for proper cellular function. By mapping these loops on an unprecedented scale, scientists are beginning to unravel how the genome’s spatial organization influences everything from development to disease progression.
The study, conducted by a team of geneticists and molecular biologists, used advanced chromosome conformation capture techniques combined with high-throughput sequencing to detect and analyze these DNA loops. The resulting map encompasses over 140,000 loops across multiple human cell types, highlighting patterns that were previously invisible to researchers. These patterns reveal how chromosomes fold within the nucleus and how looping contributes to the regulation of gene expression.
“Understanding the 3D structure of the genome is like moving from a flat map to a full topographical view,” explained Dr. Maya Lin, lead author of the study. “We can now see not just the sequence of DNA, but how it physically arranges itself to control which genes are turned on or off. This has enormous implications for biology and medicine.”
One of the most significant findings is that specific looping patterns correlate with gene activity, revealing potential mechanisms behind diseases caused by misregulated genes. Abnormal DNA looping has been linked to cancers, developmental disorders, and other genetic diseases. By understanding the normal looping architecture, researchers hope to identify how disruptions contribute to disease and explore ways to correct them.
The map also provides insight into chromosome territories—the distinct regions each chromosome occupies within the nucleus. Loops appear to play a critical role in maintaining these territories and ensuring that genes interact with the appropriate regulatory elements. This refined understanding could inform future studies on aging, epigenetics, and how environmental factors influence genome function.
Moreover, the study demonstrates the power of combining experimental biology with computational modeling. The team used sophisticated algorithms to predict and visualize loop interactions, producing a three-dimensional model of the human genome at a resolution never achieved before. These models allow scientists to explore the genome dynamically, simulating how loops form and dissolve during cell division or in response to signals.
The research community has hailed this achievement as a major milestone. While prior studies identified thousands of loops, the scale and precision of this map provide a foundation for a deeper understanding of nuclear architecture. It opens the door to new research directions, including how chromatin organization affects drug response, developmental biology, and personalized medicine.
“This is a transformative step in genomics,” said Dr. Javier Torres, a geneticist not involved in the study. “Having such a comprehensive loop map allows us to ask questions we could never tackle before, from basic biology to targeted therapies for complex diseases.”
As research continues, scientists anticipate that the DNA loop map will become an essential reference for studies in molecular biology, genetics, and biomedical research. The ability to chart the genome’s three-dimensional folding could ultimately lead to breakthroughs in diagnosing and treating diseases, as well as a better understanding of the fundamental principles that govern life at the cellular level.










Leave a Reply